Thomas Ruzicka
in cooperation with H.F. Abts and G. Michel
Genomic analysis
of drug effects
Hautklinik der Heinrich-Heine-Universität, Düsseldorf
While the completion of the human genome sequence in reliable quality is in sight, the ordering of the scattered sequences and moreover, identification of the genes within will still take some time. But even then the important question will be which of the genes from the whole transcriptome are involved in particular pathophysiological and pharmacological events. To identify candidates for these relevant genes academic biomedical research as well as biotechnology and pharmaceutical industries are forced to integrate differential gene expression profiling into their basic research, drug discovery and development strategies.
Expression profiling using mRNA differential-display (DD) offers the opportunity to identify differentially expressed genes genome-wide with high sensitivity. Since the identification is based solely on differences in mRNA abundance between the compared cell populations no sequence information of the target genes is necessary. Therefore this allows also the analysis of so far unidentified genes.
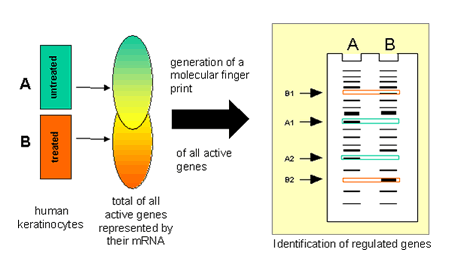
The usefulness of the approach has been demonstrated during the analysis of UV-regulated genes in human epidermal keratinocytes. Since induced and repressed genes can be detected simultaneously we were able to identify a formerly unrecognized large set of UV-repressed genes. One of these unknown sequences has been completely cloned and identified as new serine proteinase inhibitor (serpin). Serpins have not yet been connected to the UV-response. This finding emphasizes that genome-wide expression profiling by differential display does not restrict the analysis to a particular set of genes, selected based on the actual knowledge in a particular field. This advantage might also be essential when the pharmacological modes of action should be dissected. The genes which are regulated in a cell by a pharmacological substance form the molecular basis of the desired curative effect as well as the unwanted site effects. The knowledge of these genes will allow to elucidate the mechanisms underlying both actions.
By a restricted differential display PCR analysis several genes which are regulated in human keratinocytes by antihistaminic substances have been identified. The majority of the isolated sequences have been so far only described as ESTs or anonymous sequence information from the human genome project. Although this documents the enormous increase in sequence data in the databases due to the human genome project, there were still completely new sequences identified. Among the known sequences the cytoplasmic protein a-catenin was identified. This protein plays a crucial role in E-cadherin mediated cell-cell adhesion by binding E-cadherin to the cytoskeleton via beta-catenin or gamma-catenin and actin. These results implicate that antihistaminica may influence epithelial integrity by modulating the expression of relevant genes like the catenins.
In summary expression profiling will identify not only new targets for future drug development but also allow the identification of the molecular basis of drug action and unwanted side effects. This information will help to optimize current drugs and to individualize therapy based on pharmacogenomic analysis.




| Copyright © 2000 -
2024 Institute for Dermopharmacy GmbH webmaster@gd-online.de |
Impressum Haftungsausschluss |